|
|
| -Genomic Data for CSL |
Links |
- Protein
- CSL
|
- UniPROBE Accession Number
- UP00484
|
- Species
- Homo sapiens
|
- Domain
- LAG1-DNAbind
|
- Swiss-Prot
- Q06330
|
- Name and Synonyms
- Name: recombination signal binding protein for immunoglobulin kappa J region.
Synonyms:SUH; csl; CBF1; KBF2; RBPJ; RBP-J; RBPJK; IGKJRB; RBPSUH; IGKJRB1; MGC61669
|
- IHOP
- 89405
|
- NCBI RefSeq
- NP_976028.1
|
- Description
- Transcriptional regulator that plays a central role in Notch signaling, a signaling pathway involved in cell-cell communication that regulates a broad spectrum of cell-fate. determinations. Acts as a transcriptional repressor when it is not associated with Notch proteins. When associated with some Notch protein, it acts as a transcriptional activator that activates transcriptio
|
- JASPAR
-
None Available
|
| -PBM Motif Data for GST-CSL-NOTCH1 |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
CGTGGGAA E.S. 0.498
TTCCCACG E.S. 0.498
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-
|
- PWM
- Save
|
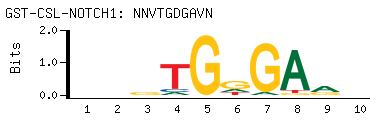
- Consensus
-
NNVTGDGAVN
NBTCHCABNN
|
- Downloads
-
Download the zip
of all GST-CSL-NOTCH1 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
| -PBM Motif Data for GST-CSL_Fig2 |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
CGTGGGAA E.S. 0.498
TTCCCACG E.S. 0.498
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-
|
- PWM
- Save
|
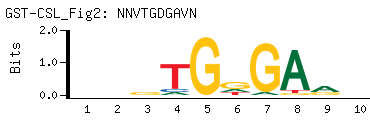
- Consensus
-
NNVTGDGAVN
NBTCHCABNN
|
- Downloads
-
Download the zip
of all GST-CSL_Fig2 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
| -PBM Motif Data for GST-CSL_Fig4 |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
CGTGGGAA E.S. 0.499
TTCCCACG E.S. 0.499
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-

|
- PWM
- Save
|
- Consensus
-
NNVTGDGARN
NYTCHCABNN
|
- Downloads
-
Download the zip
of all GST-CSL_Fig4 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
| -PBM Motif Data for GST-NOTCH1_CSL-6His |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
C.TGGGAAC E.S. 0.495
GTTCCCA.G E.S. 0.495
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-
|
- PWM
- Save
|
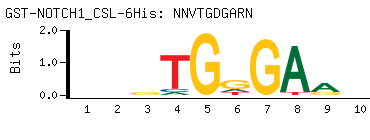
- Consensus
-
NNVTGDGARN
NYTCHCABNN
|
- Downloads
-
Download the zip
of all GST-NOTCH1_CSL-6His PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
| -PBM Motif Data for GST-NOTCH1_CSL-6His_MAML1 |
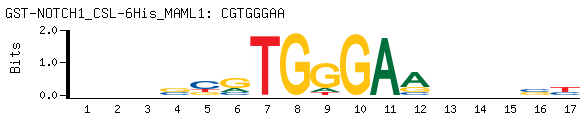
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-
|
- PWM
- Save
|
- Top Kmer
-
CGTGGGAA E.S. 0.497
TTCCCACG E.S. 0.497
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-

|
- PWM
- Save
|
- Consensus
-
NNVTGDGARN
NYTCHCABNN
|
- Downloads
-
Download the zip
of all GST-NOTCH1_CSL-6His_MAML1 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
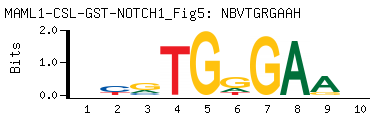
| -PBM Motif Data for MAML1-CSL-GST-NOTCH1_Fig5 |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
C.TGGGAA.C E.S. 0.493
G.TTCCCA.G E.S. 0.493
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-
|
- PWM
- Save
|
- Consensus
-
NBVTGRGAAH
DTTCYCABVN
|
- Downloads
-
Download the zip
of all MAML1-CSL-GST-NOTCH1_Fig5 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
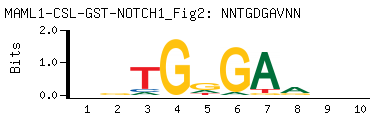
| -PBM Motif Data for MAML1-CSL-GST-NOTCH1_Fig2 |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
C.TGGGAAC E.S. 0.498
GTTCCCA.G E.S. 0.498
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-
|
- PWM
- Save
|
- Consensus
-
NNTGDGAVNN
NNBTCHCANN
|
- Downloads
-
Download the zip
of all MAML1-CSL-GST-NOTCH1_Fig2 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
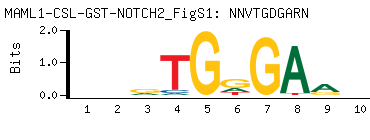
| -PBM Motif Data for MAML1-CSL-GST-NOTCH2_FigS1 |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
C..TGGGAAC E.S. 0.495
GTTCCCA..G E.S. 0.495
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-
|
- PWM
- Save
|
- Consensus
-
NNVTGDGARN
NYTCHCABNN
|
- Downloads
-
Download the zip
of all MAML1-CSL-GST-NOTCH2_FigS1 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
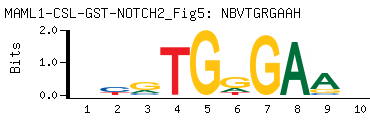
| -PBM Motif Data for MAML1-CSL-GST-NOTCH2_Fig5 |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
CC.TGGGAA E.S. 0.492
TTCCCA.GG E.S. 0.492
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-
|
- PWM
- Save
|
- Consensus
-
NBVTGRGAAH
DTTCYCABVN
|
- Downloads
-
Download the zip
of all MAML1-CSL-GST-NOTCH2_Fig5 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
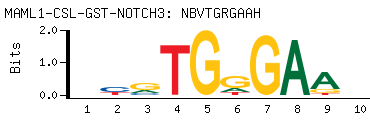
| -PBM Motif Data for MAML1-CSL-GST-NOTCH3 |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
CC.TGGGAA E.S. 0.494
TTCCCA.GG E.S. 0.494
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-
|
- PWM
- Save
|
- Consensus
-
NBVTGRGAAH
DTTCYCABVN
|
- Downloads
-
Download the zip
of all MAML1-CSL-GST-NOTCH3 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
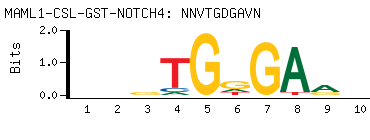
| -PBM Motif Data for MAML1-CSL-GST-NOTCH4 |
- PBM-Derived DNA Binding Site Motif (Seed-And-Wobble)
-

|
- PWM
- Save
|
- Top Kmer
-
C..TGGGAAC E.S. 0.496
GTTCCCA..G E.S. 0.496
|
- PBM-Derived DNA Binding Site Motif (BEEML-PBM)
-
|
- PWM
- Save
|
- Consensus
-
NNVTGDGAVN
NBTCHCABNN
|
- Downloads
-
Download the zip
of all MAML1-CSL-GST-NOTCH4 PBM files
or view the downloads directory
for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
| -CSL Datasets |
| Complex Name |
Save |
View |
| GST-CSL-NOTCH1 |

|

|
| Run name |
PBM version |
Save |
View |
| GST-CSL-NOTCH1 | AMADID # 015681 |

|

|
| Complex Name |
Save |
View |
| GST-CSL |

|

|
| Run name |
PBM version |
Save |
View |
| GST-CSL_Fig2 | AMADID # 015681 |

|

|
| GST-CSL_Fig4 | AMADID # 015681 |

|

|
| Complex Name |
Save |
View |
| GST-NOTCH1_CSL-6His |

|

|
| Run name |
PBM version |
Save |
View |
| GST-NOTCH1_CSL-6His | AMADID # 015681 |

|

|
| Complex Name |
Save |
View |
| GST-NOTCH1_CSL-6His_MAML1 |

|

|
| Run name |
PBM version |
Save |
View |
| GST-NOTCH1_CSL-6His_MAML1 | AMADID # 015681 |

|

|
| Complex Name |
Save |
View |
| MAML1-CSL-GST-NOTCH1 |

|

|
| Run name |
PBM version |
Save |
View |
| MAML1-CSL-GST-NOTCH1_Fig5 | AMADID # 015681 |

|

|
| MAML1-CSL-GST-NOTCH1_Fig2 | AMADID # 015681 |

|

|
| Complex Name |
Save |
View |
| MAML1-CSL-GST-NOTCH2 |

|

|
| Run name |
PBM version |
Save |
View |
| MAML1-CSL-GST-NOTCH2_FigS1 | AMADID # 015681 |

|

|
| MAML1-CSL-GST-NOTCH2_Fig5 | AMADID # 015681 |

|

|
| Complex Name |
Save |
View |
| MAML1-CSL-GST-NOTCH3 |

|

|
| Run name |
PBM version |
Save |
View |
| MAML1-CSL-GST-NOTCH3 | AMADID # 015681 |

|

|
| Complex Name |
Save |
View |
| MAML1-CSL-GST-NOTCH4 |

|

|
| Run name |
PBM version |
Save |
View |
| MAML1-CSL-GST-NOTCH4 | AMADID # 015681 |

|

|
| Gene |
Clone Type |
Last Modified |
Save |
View |
| CSL |
Not full length |
2014-08-13 |
N/A
|
N/A
|
- Insert Sequence
-
1 ERPPPKRLTR EAMRNYLKER GDQTVLILHA KVAQKSYGNE KRFFCPPPCV
51 YLMGSGWKKK KEQMERDGCS EQESQPCAFI GIGNSDQEMQ QLNLEGKNYC
101 TAKTLYISDS DKRKHFMLSV KMFYGNSDDI GV FLSKRIK VISKPSKKKQ
151 SLKNADLCIA SGTKVALFNR LRSQTVSTRY LHVEGGNFHA SSQQWGAFFI
201 HLLDDDESEG EEFTVRDGYI HYGQTVKLVC SVTGMALPRL IIRKVDKQTA
251 LLDADDPVSQ LHKCAFYLKD TER MYLCLS QERIIQFQAT PCPKEPNKEM
301 INDGASWTII STDKAEYTFY EGMGPVLAPV TPVPVVESLQ LNGGGDVAML
351 ELTGQNFTPN LRVWFGDVEA ETMYRCGESM LCVVPDISAF REGWRWVRQP
401 VQVPVTLVRN DGIIYSTSLT FTYTPEPGP
|
- References
-
Data are analyzed by array design with AMADID # 015681 in Del Bianco et al., PLoS ONE 2010
|
|