|
|
| -Genomic Data |
Links |
- Protein
- TBP
|
- UniPROBE Accession Number
- UP00029
|
- Species
- Homo sapiens
|
- Domain
- TBP
|
- Uniprot
-
P20226
|
- Name and Synonyms
-
TATA-box-binding protein
TATA-box factor, TATA sequence-binding protein, Transcription initiation factor TFIID TBP subunit, GTF2D; GTF2D1, HDL4, MGC117320, MGC126054, MGC126055, SCA17, TATA-binding factor, TATA-box-binding protein, TF2D, TFIID
|
- IHOP
-
92558
|
- RefSeq
-
NP_003185
|
- Description
- Initiation of transcription by RNA polymerase II requires the activities of more than 70 polypeptides. The protein that coordinates these activities is transcription factor IID (TFIID), which binds to the core promoter to position the polymerase properly, serves as the scaffold for assembly of the remainder of the transcription complex, and acts as a channel for regulatory signals. TFIID is composed of the TATA-binding protein (TBP) and a group of evolutionarily conserved proteins known as TBP-associated factors or TAFs. TAFs may participate in basal transcription, serve as coactivators, function in promoter recognition or modify general transcription factors (GTFs) to facilitate complex assembly and transcription initiation. This gene encodes TBP, the TATA-binding protein. A distinctive feature of TBP is a long string of glutamines in the N-terminus. This region of the protein modulates the DNA binding activity of the C terminus, and modulation of DNA binding affects the rate of transcription complex formation and initiation of transcription. The number of CAG repeats encoding the polyglutamine tract is usually 32-39, and expansion of the number of repeats increases the length of the polyglutamine string and is associated with spinocerebellar ataxia 17, a neurodegenerative disorder classified as a polyglutamine disease. Two transcript variants encoding different isoforms have been found for this gene.
|
- JASPAR
-
MA0108
|
| -PBM Motif Data for TBP |
- Primary Motif (Seed-And-Wobble)
-

|
- Primary PWM
-
View
Save
|
- Top Kmer
-
TATATATA E.S. 0.496
TATATATA E.S. 0.496
|
- Secondary Motif (Seed-And-Wobble)
-

|
- Secondary PWM
-
View
Save
|
- Top Kmer
-
ATATAAG.G E.S. 0.433
C.CTTATAT E.S. 0.433
|
-
Motif (BEEML-PBM) (AMADID # 015681)
(clone pTH0781)
-
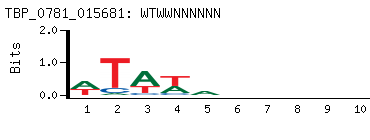
|
- PWM
-
View
|
- Consensus
-
WTWWNNNNNN
NNNNNNWWAW
|
-
Motif (BEEML-PBM) (AMADID # 016060) (clone pTH0781)
-

|
- PWM
-
View
|
- Consensus
-
NNNNBVCCYS
SRGGBVNNNN
|
- Downloads
-
Download the
zip
of all TBP PBM files
or view the
downloads directory
for individual files.
|
- Link to TFBSshape
-
This link will take you to the corresponding entry for TBP
in TFBSshape,
a database which provides information about the shape of the DNA at transcription factor binding sites.
http://rohslab.cmb.usc.edu/TFBSshape/?tfid=&geneID=29&sourceDB=uniprobe
|
| -TBP Datasets |
| Clone ID |
Expression |
Array Version |
Expression Protocol |
Clone Type |
Last Modified |
Save |
View |
|
pTH781 |
#1 |
AMADID #016060 |
Commercially Purified Protein |
DNA binding domain only |
2014-08-21 |

|

|
| Clone ID |
Expression |
Array Version |
Expression Protocol |
Clone Type |
Last Modified |
Save |
View |
|
pTH781 |
#1 |
AMADID #015681 |
Commercially Purified Protein |
DNA binding domain only |
2014-08-21 |

|

|
- Clone pTH781 insert sequence
- 1 MTPITPATPA SESSGIVPQL QNIVSTVNLG CKLDLKTIAL RARNAEYNPK
51 RFAAVIMRIR EPRTTALIFS SGKMVCTGAK SEEQSRLAAR KYARVVQKLG
101 FPAKFLDFKI QNMVG
- DNA Binding Domains (Pfam) for clone 781
- Not Available
|
- References
-
Data are analyzed by array designs with AMADID #s 016060, 015681 in Badis et al., Science 2009
|
|