|
|
| -Genomic Data for Zif268 |
Links |
- Protein
|
- UniPROBE Accession Number
- UP00400
|
- Species
- Mus musculus
|
- Domain
- zf-C2H2
|
- Swiss-Prot
- P08046
|
- Name and Synonyms
- Name: Transcription factor Zif268.
Synonyms:early growth response 1, A530045N19Rik, Early growth response protein 1, egr, Egr-1, EGR-1, ETR103, Krox-1, Krox24, Krox-24, Krox-24 protein, Nerve growth factor-induced protein A, NGF1-A, NGFIA, NGFI-A, TIS8, Transcription factor Zif268, Zenk, Zfp-6
|
- IHOP
- 120755
|
- NCBI RefSeq
- NP_031939
|
- Description
- None Available
|
- JASPAR
-
N/A
|
| -PBM Motif Data for Zif268 |
- PBM-Derived DNA Binding Site Motif
(Seed-And-Wobble)
-

|
- PWM
-
Save
|
- Top Kmer
-
C.CCCACGC E.S. 0.497
GCGTGGG.G E.S. 0.497
|
- PBM-Derived DNA Binding Site Motif
(BEEML-PBM) (AMADID # 015681)
-
|
- PWM
-
Save
|
- Consensus
-
NHDYMYTHDH
DHDARKRHDN
|
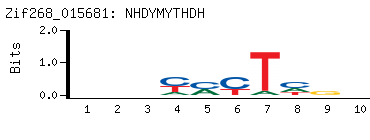
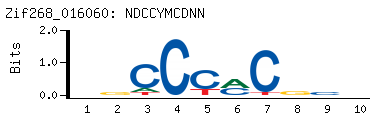
- PBM-Derived DNA Binding Site Motif
(BEEML-PBM) (AMADID # 016060)
-
|
- PWM
-
Save
|
- Consensus
-
NDCCYMCDNN
NNHGKRGGHN
|
- Downloads
-
Download the
zip
of all Zif268 PBM files
or view the downloads directory for individual files.
Please note that the files which include probe sequence data are protected by an academic research use license,
which can be viewed here.
|
- Link to TFBSshape
-
This link will take you to the corresponding entry for Zif268
in TFBSshape,
a database which provides information about the shape of the DNA at transcription factor binding sites.
http://rohslab.cmb.usc.edu/TFBSshape/?tfid=&geneID=400&sourceDB=uniprobe
|
| -Zif268 Datasets |
| Gene |
Clone Type |
Last Modified |
Save |
View |
|
Zif268 |
Full length clone |
2014-07-16 |

|

|
- Zif268 Insert Sequence
-
1 MAAAKAEMQL MSPLQISDPF GSFPHSPTMD NYPKLEEMML LSNGAPQFLG
51 AAGTPEGSGG NSSSSTSSGG GGGGGSNSGS SAFNPQGEPS EQPYEHLTTE
101 SFSDIALNNE KAMVETSYPS QTTRLPPITY TGRFSLEPAP NSGNTLWPEP
151 LFSLVSGLVS MTNPPTSSSS APSPAASSSS SASQSPPLSC AVPSNDSSPI
201 YSAAPTFPTP NTDIFPEPQS QAFPGSAGTA LQYPPPAYPA TKGGFQVPMI
251 PDYLFPQQQG DLSLGTPDQK PFQGLENRTQ QPSLTPLSTI KAFATQSGSQ
301 DLKALNTTYQ SQLIKPSRMR KYPNRPSKTP PHERPYACPV ESCDRRFSRS
351 DELTRHIRIH TGQKPFQCRI CMRNFSRSDH LTTHIRTHTG EKPFACDICG
401 RKFARSDERK RHTKIHLRQK DKKADKSVVA SPAASSLSSY PSPVATSYPS
451 PATTSFPSPV PTSYSSPGSS TYPSPAHSGF PSPSVATTFA SVPPAFPTQV
501 SSFPSAGVSS SFSTSTGLSD MTATFSPRTI EIC
|
- References
-
Data are analyzed by array design with AMADID # in Berger et al., Nat Biotech 2006
|
|