
In the event that UniPROBE does not contain PBM data for the specific query protein, this search tool finds PBM data of other, similar proteins. The tool calls directly to a locally-hosted instance of the NCBI blastp program (Altschul S et al., Nucleic Acids Res 1997) from the Blast suite (v. 2.2.23).
The output data from blastp, which describes successful matches, are then
displayed in the 'Browse' page. Provided are:
* Name of the matched target protein.
* Offset within the query protein for which the alignment starts.
* Raw score, a measure of the alignment similarity.
* E-value, or the number of expected matches if the database were comprised of randomly-generated protein sequences.
* Sequence of the matched target protein from UniPROBE, with the entire span of the alignment highlighted.
* Publication for the matched target protein.
* Link to the details page of the matched target protein.
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A. Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402.
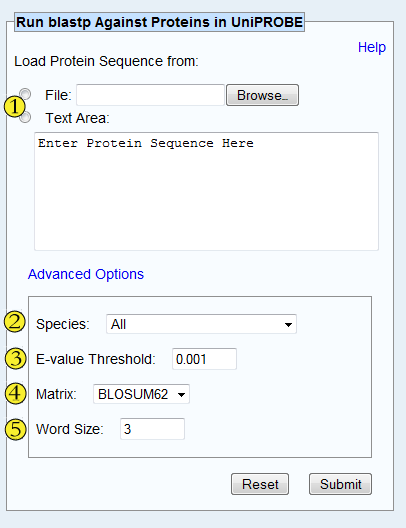
 Submit the FASTA data either by uploading a file or by insering the
text directly into the Text Area. For example:
Submit the FASTA data either by uploading a file or by insering the
text directly into the Text Area. For example:
>Pbf1 MTLPKLSSVSVSSGHVSANSHGFSILSKHPHPNNLVHSHSLSHTNAKSHLPISSTSTKEN STNKEEAESLKKNNPSSWDPSDDIKLRHLKEIKNLGWKEIAHHFPNRTPNACQFRWRRLK SGNLKSNKTAVIDINKLFGVYATGDATPSAGTPSAEEAVKEEAVEDEDITAGSSAIEDSP PDFKPLVKPKYMDRKLITQRSTSTFSDHEPQHTKPRKLFVKPRSFSHSITTNTPNVKTAQ QTNLSLYNTTSAKTNKAVNSNDYENIGLVPKIIIRSRRNSFIPSTQIPHSTTKTRKNSHS VISSRRSSFNMMHSRRSSFNSHAPTEPISRRASLVVSPYMSPRRLSTSQSVHYHPQHQYY LNPIASPNCKTDHANDKITHTRTFLDMQKFANKHPWSREDDEVLLNNTKDKQNHLSPLEI SIVLPNNRSELEIQQRMDYLKRKGRVSGFHTNEGCKDEEEEDDIDPLHKENGINTPSQQS QNYGMLEAKHDNPKSSELSSMTSANDIRNEQDELPGINSIFKNIF >Apterous MGVCTEERPVMHWQQSARFLGPGAREKSPTPPVAHQGSNQCGSAAGANNN HPLFRACSSSSCPDICDHSTKPFGNAYGTESFRSYETADRATFEDSAAKF SISRSRTDCTEVSDETTSGISFKTEPFGPPSSPESTSDSKITRNLDDCSG CGRQIQDRFYLSAVEKRWHASCLQCYACRQPLERESSCYSRDGNIYCKND YYSFFGTRRCSRCLASISSNELVMRARNLVFHVNCFCCTVCHTPLTKGDQ YGIIDALIYCRTHYSIAREGDTASSSMSATYPYSAQFGSPHNDSSSPHSD PSRSIVPTGIFVPASHVINGLPQPARQKGRPRKRKPKDIEAFTANIDLNT EYVDFGRGSHLSSSSRTKRMRTSFKHHQLRTMKSYFAINHNPDAKDLKQL SQKTGLPKRVLQVWFQNARAKWRRMMMKQDGSGLLEKGEGALDLDSISVH SPTSFILGGPNSTPPLNLD
As an alternative to the FASTA format, the user may provide a file (or text in the Text Area) where each line contains a single protein sequence. Up to 20 amino acid sequences can be entered in batch, each with length no greater than 10,000 residues. Upon submission, the tool will then label the input as sequences 1 through n, where n is the number of lines entered (or the FASTA tag for that sequence). Acceptable amino acid codes are the same dictated by blastp, namely:
A alanine P proline
B aspartate or asparagine Q glutamine
C cystine R arginine
D aspartate S serine
E glutamate T threonine
F phenylalanine U selenocysteine
G glycine V valine
H histidine W tryptophan
I isoleucine Y tyrosine
K lysine Z glutamate or glutamine
L leucine X any
M methionine * translation stop
N asparagine - gap of indeterminate length
 Limit your search by species, or choose 'All' to include all species in the search.
Limit your search by species, or choose 'All' to include all species in the search.
 Only include results with the specified E-value or lower. The E-value
is defined by blastp as the number of expected matches if the database
were comprised of randomly-generated protein sequences.
Only include results with the specified E-value or lower. The E-value
is defined by blastp as the number of expected matches if the database
were comprised of randomly-generated protein sequences.
 Pick the amino acid similarity matrix to use for the blastp search.
Pick the amino acid similarity matrix to use for the blastp search.
 Word size for blastp's word finder algorithm. Choose a size >= 2.
Word size for blastp's word finder algorithm. Choose a size >= 2.