In order to automate the motif searches, we developed a software package, termed MultiFinder, that performs automated motif searching using four different profile-based motif finders, including AlignACE, MDscan, BioProspector and MEME. We anticipated that using all four of these motif finders might allow the user to combine the strengths of their different algorithms.
The integration of the results from multiple motif finding tools identifies and ranks highly more known and novel motifs than does the use of just one of these tools. In addition, we believe that our simultaneous enrichment strategies helped to identify likely human cis regulatory elements. A number of the discovered motifs may correspond to novel binding site motifs for as yet uncharacterized tissue-specific TFs. We expect this strategy to be useful for identifying motifs in other metazoan genomes.

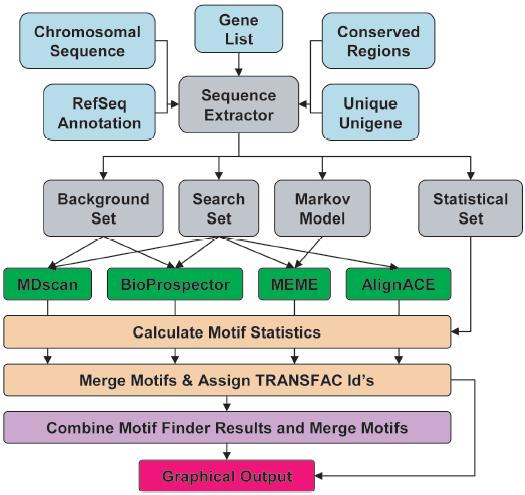
The sequence and support files are input files (aqua boxes) required by the sequence extraction script SequenceExtractor.pl to generate a number of input files (gray boxes) required by the motif finders MDscan, BioProspector, MEME, and AlignACE (green boxes). Statistics are generated for the motifs, similar motifs are merged and TRANSFAC IDs are assigned (tan boxes). Motifs from the previous step are combined and similar motifs are merged (purple box). Graphical output of the combined results (purple box) from all four motif finders is generated for each scoring function (red box).