Compact, universal DNA microarrays
to comprehensively determine transcription-factor binding site specificities.
This website supports
Berger, Philippakis, et al.
Nature Biotechnology, Epub 2006 Sept 24
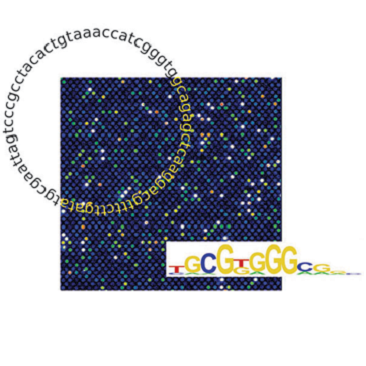
We have created a novel, maximally compact, synthetic DNA sequence design for
protein binding microarrays (PBMs) that represents
all possible DNA sequence variants of a given length k in an overlapping
fashion on a single slide. We constructed such ‘all 10-mer’ microarrays by converting high-density single-stranded
Agilent oligonucleotide arrays to double-stranded DNA
arrays via on-array primer extension. Using these microarrays,
we comprehensively determined the binding specificities over a full range of
affinities of several TFs of diverse structural
classes from yeast, worm, mouse, and human. We also developed a novel
computational method to construct TF binding site motifs that takes full
advantage of the unbiased sequence representation on these arrays, using all
features (rather than only those above an arbitrary cutoff) to determine the
optimal motif without any prior knowledge. As advances in microarray printing
technology permit increased feature densities and feature lengths, our
universal design will enable the complete coverage of even longer binding
sites. This unbiased, comprehensive coverage of all k-mers permits interrogation of binding site preferences,
including nucleotide interdependencies, at unprecedented resolution.
Below are the Supplementary data files that accompany this manuscript.
For the normalized signal intensities and DNA probe sequences of our two separate
Agilent ‘all 10-mer’ universal microarrays, please click here.
Survey of binding sites in the JASPAR database.
Reproducibility of Cy3 dUTP signal intensities.
Correlation of PBM signal intensities with affinities.
Effects of binding site position and orientation on PBM signal.
Comparison of median signal intensities for 28 Zif268 variants for fixed versus variable position, orientation, and flanking sequence.
Correspondence between median signal intensities for 7-mers on distinct de Bruijn sequences.
Biacore measurements supporting interdependence between the first two positions of the Cbf1 binding site.
Minimum number of unique features on an array for different values of k.
Dependence of Cy3 dUTP incorporation upon sequence context.
Questions? Mike Berger, Anthony Philippakis, Martha Bulyk.